Methodology
Increasingly, the effectiveness of adjuvant chemotherapy agents for breast cancer has
been related to changes in the genomic profile of tumors. We investigated correspondence between growth
inhibitory concentrations of paclitaxel and gemcitabine (GI50) and gene copy number, mutation, and expression
first in breast cancer cell lines and then in patients. Genes encoding direct targets of these drugs,
metabolizing enzymes, transporters, and those previously associated with chemoresistance to paclitaxel
(n=31 genes) or gemcitabine (n=18) were analyzed. A multi-factorial, principal component analysis (MFA)
indicated expression was the strongest indicator of sensitivity for paclitaxel, and copy number and expression were informative for
gemcitabine. The factors were combined using support vector machines (SVM). Expression of 15 genes (ABCC10,
BCL2, BCL2L1, BIRC5, BMF, FGF2, FN1, MAP4, MAPT, NKFB2, SLCO1B3, TLR6, TMEM243, TWIST1, and CSAG2) predicted
cell line sensitivity to paclitaxel with 82% accuracy. Copy number profiles of 3 genes (ABCC10, NT5C, TYMS)
together with expression of 7 genes (ABCB1, ABCC10, CMPK1, DCTD, NME1, RRM1, RRM2B), predicted gemcitabine
response with 85% accuracy. Expression and copy number studies of two independent sets of patients with known
responses were then analyzed with these models. These included tumour blocks from 21 patients that were
treated with both paclitaxel and gemcitabine, and 319 patients on paclitaxel and anthracycline therapy.
A new paclitaxel SVM was derived from an 11-gene subset since data for 4 of the original genes was unavailable.
The accuracy of this SVM was similar in cell lines and tumour blocks (70-71%). The gemcitabine SVM exhibited 62%
prediction accuracy for the tumour blocks due to the presence of samples with poor nucleic acid integrity. Nevertheless,
the paclitaxel SVM predicted sensitivity in 84% of patients with no or minimal residual disease.
Growth inhibition (GI50) values for paclitaxel and gemcitabine, as well as copy number and gene expression data
were obtained from the supplementary data of Daemen et al. (2013). The data was used to train SVMs on a
set of cell lines for paclitaxel and gemcitabine.
The SVM calculator requires data comparable to the training set for classification. Log2 normalized gene expression data were derived from Affymetrix Gene Chip Human Exon 1.0 ST arrays. Array data from Exon 1.0 ST and U133 arrays is comparable to this dataset (after log2 normalization) and can be entered into the SVM calculator as is. Copy number data must be entered as an integer.
qRT-PCR data was also used as a testing set. FFPE qRT-PCR data was normalized against ACTB, B2M, and GAPDH according to the equation:
expression value = 2-ΔCt = 2-(Ct GOI - average Ct of standards)
That data must be binned to be comparable to the test set. Input the data and activate the binning option at the bottom of the form and the data will automatically be binned into pre-existing qRT-PCR bins.
The SVM calculator requires data comparable to the training set for classification. Log2 normalized gene expression data were derived from Affymetrix Gene Chip Human Exon 1.0 ST arrays. Array data from Exon 1.0 ST and U133 arrays is comparable to this dataset (after log2 normalization) and can be entered into the SVM calculator as is. Copy number data must be entered as an integer.
qRT-PCR data was also used as a testing set. FFPE qRT-PCR data was normalized against ACTB, B2M, and GAPDH according to the equation:
expression value = 2-ΔCt = 2-(Ct GOI - average Ct of standards)
That data must be binned to be comparable to the test set. Input the data and activate the binning option at the bottom of the form and the data will automatically be binned into pre-existing qRT-PCR bins.
An SVM model was trained for paclitaxel and gemcitabine: using expression values binned into 10 categories.
Binning was performed because amplifiable RNA template concentration in FFPE blocks is not known precisely,
because it is subject to long term degradation and reactivity. Equal width binning was performed, where the
date is divided into 10 intervals of equal size. If a value is greater than the previous bin, and less than or equal
to the current bin, it falls into that bin.
The binning tables are show below. ABCC10 is in the gemcitabine and paclitaxel set so does not repeat between tables - i.e. it is only in the paclitaxel table.
The binning tables are show below. ABCC10 is in the gemcitabine and paclitaxel set so does not repeat between tables - i.e. it is only in the paclitaxel table.
Threshold Gene Expression Value for Binning (*10-4)
Paclitaxel Genes
| Bin No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| ABCC10 | 1.03 | 2.37 | 3.29 | 4.49 | 5.73 | 7.68 | 11.36 | 15.85 | 29.48 | 60.98 |
| BCL2 | 1.52 | 3.29 | 5.00 | 7.20 | 14.11 | 30.48 | 52.26 | 82.10 | 133.4 | 391.1 |
| BCL2L1 | 33.29 | 86.96 | 152.3 | 171.2 | 246.8 | 336.2 | 359.6 | 496.7 | 658.0 | 1897 |
| BIRC5 | 3.04 | 4.52 | 15.55 | 51.02 | 11.73 | 26.52 | 11.73 | 493.3 | 1704.6 | 3270.2 |
| FGF2 | 3.29 | 4.68 | 8.30 | 14.51 | 37.80 | 66.40 | 93.00 | 219.1 | 355.15 | 557.1 |
| FN1 | 97.79 | 41.79 | 67.56 | 95.13 | 1070.2 | 1290.4 | 1453.2 | 1526.7 | 1920.9 | 2639.7 |
| MAP4 | 58.50 | 18.82 | 25.05 | 32.70 | 37.86 | 44.19 | 51.66 | 58.43 | 69.11 | 79.32 |
| MAPT | 1.49 | 3.29 | 5.88 | 8.88 | 15.07 | 24.03 | 37.61 | 46.65 | 153.7 | 291.7 |
| NFKB2 | 5.41 | 15.09 | 51.68 | 127.3 | 178.7 | 225.5 | 249.6 | 414.8 | 540.5 | 830.0 |
| TLR6 | 0.847 | 1.44 | 3.29 | 4.49 | 7.76 | 11.48 | 16.98 | 35.35 | 116.5 | 403.6 |
| TMEM243 | 1.84 | 4.57 | 7.68 | 15.05 | 19.42 | 25.30 | 39.73 | 77.71 | 119.1 | 239.7 |
Threshold Gene Expression Value for Binning (*10-4)
Gemcitabine Genes
| Bin No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| CMPK1 | 720.8 | 959.0 | 1234.5 | 1342.9 | 1451.2 | 1580.4 | 1855.7 | 2348.9 | 2811.6 | 3855.0 |
| DCTD | 67.08 | 124.1 | 238.6 | 308.9 | 329.3 | 419.3 | 448.2 | 503.6 | 588.3 | 771.5 |
| NME1 | 3.30 | 5.24 | 12.80 | 20.60 | 35.02 | 72.24 | 114.5 | 139.1 | 322.0 | 1644 |
| RRM1 | 0.746 | 2.00 | 3.43 | 5.24 | 7.35 | 10.15 | 16.16 | 29.26 | 42.85 | 184.6 |
| RRM2B | 6.12 | 9.48 | 19.54 | 39.70 | 58.84 | 115.3 | 152.0 | 214.4 | 277.2 | 1001 |
The SVM Calculator will return a classification for the sample ("sensitive" or "resistant").
It will also return the classification score - the distance from the SVM hyperplane that distinguishes
sensitive or resistant data. If you have selected the binning function, it will return the results of
the binning on the next page.
The box and whisker plot below will aid in interpretation of the SVM score. Patients from Hatzis et. al. (2011) with a low residual disease burden were classified by our expression signature to predict pathological complete response (pCR) or recurrent disease (RD). The bow and whisker plot demonstrates the score for each class: RD and pCR misclassified as resistant (scores above 0) and pCR and RD misclassified as sensitive (scores below 0). A "sensitive" result is more likely to have been misclassified when it returns a score close to 0 (the position of the hyeperplane). However, this is not the case for "resistant" results, so caution must be taken here.
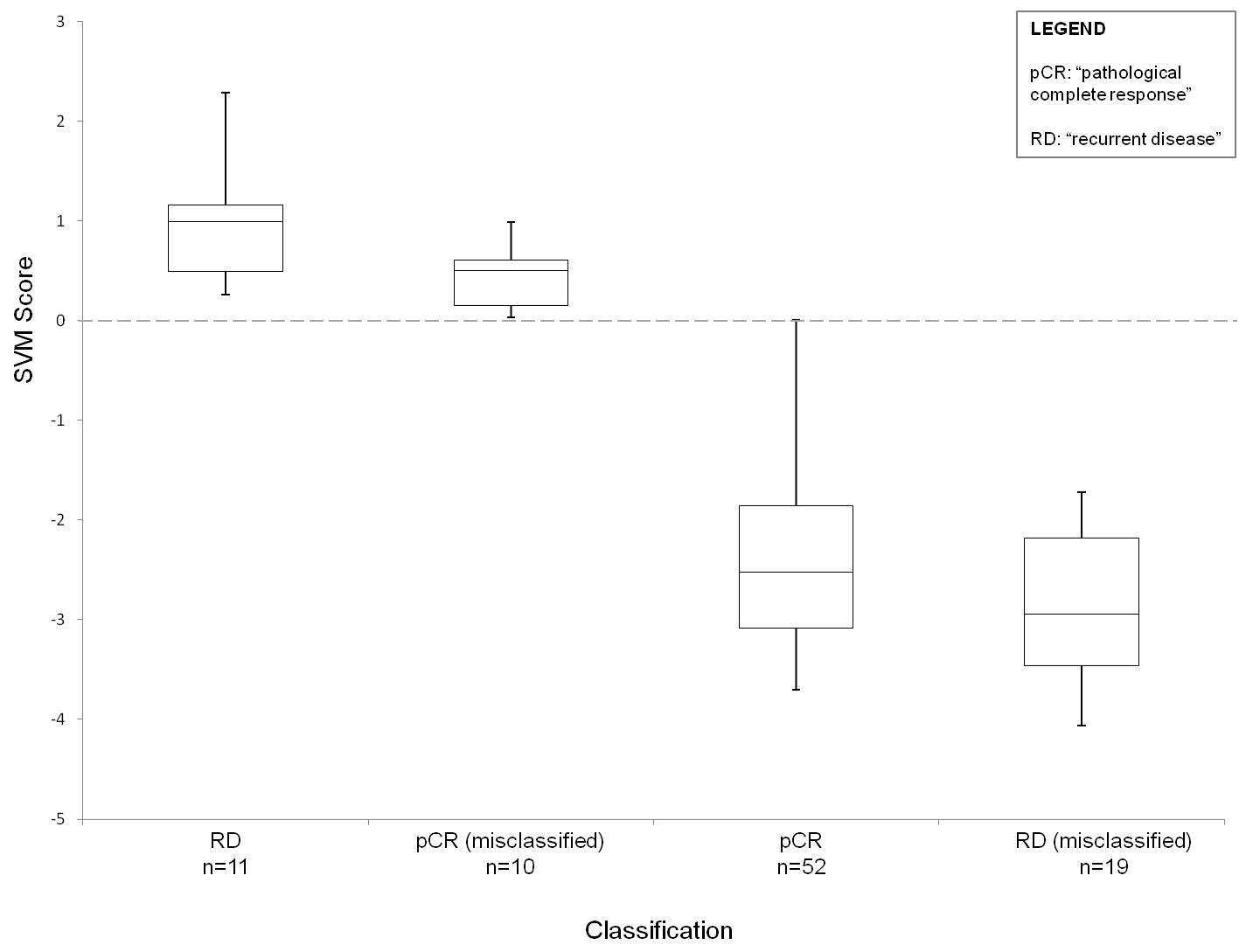
The box and whisker plot below will aid in interpretation of the SVM score. Patients from Hatzis et. al. (2011) with a low residual disease burden were classified by our expression signature to predict pathological complete response (pCR) or recurrent disease (RD). The bow and whisker plot demonstrates the score for each class: RD and pCR misclassified as resistant (scores above 0) and pCR and RD misclassified as sensitive (scores below 0). A "sensitive" result is more likely to have been misclassified when it returns a score close to 0 (the position of the hyeperplane). However, this is not the case for "resistant" results, so caution must be taken here.
